I'm pursuing an MSc in Health Data Science and my dissertation focuses on implementing a quantum principal component analysis (qPCA) algorithm using IBM quantum hardware. My goal is to compare classical and a quantum PCA methods, applying them to fMRI data represented as covariance matrices. In my experiments, I plan to scale from small matrices (e.g., 2×2 or 4×4) to larger ones (e.g., 12×12).
THESE ARE TWO PAPERS IM CURRENTLY USING TO TRY DEVELOP MY METHODS:
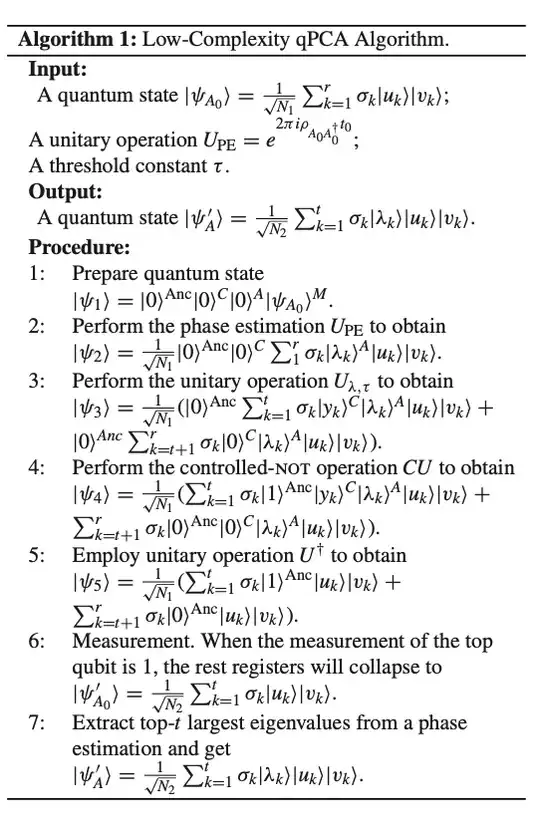
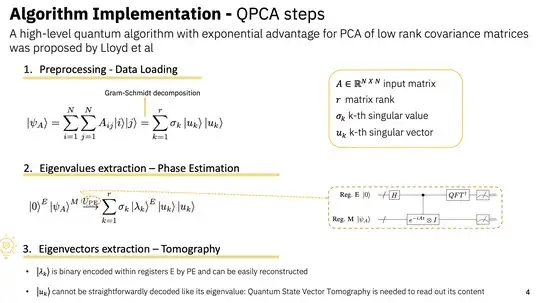
A LOW COMPLEXITY QPCA : https://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=9669030
END TO END APPROACH: https://ieeexplore.ieee.org/stamp/stamp.jsp?tp=&arnumber=10313821.
My questions are as follows:
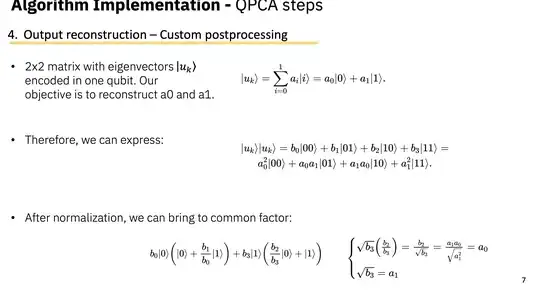
Data Encoding & Initialization: If I implement quantum singular value thresholding (QSVT) using block encoding, what data encoding and state initialization techniques would be the most versatile and suitable for my application, especially when comparing against classical PCA? and possibly adding a different quantum algorithm but keeping the encoding the same?
Scalability: What strategies can be used to manage scalability and computational complexity when transitioning from smaller matrices (2×2 or 4×4) to a 12×12 matrix, particularly in the context of fMRI data?
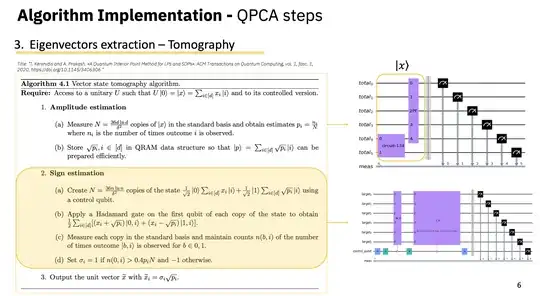
Method Selection: Considering my objectives, should I focus on QSVT or explore quantum state vector tomography for the state preparation part of the algorithm? What are the trade-offs between these methods regarding accuracy, circuit depth, and overall feasibility on current hardware?