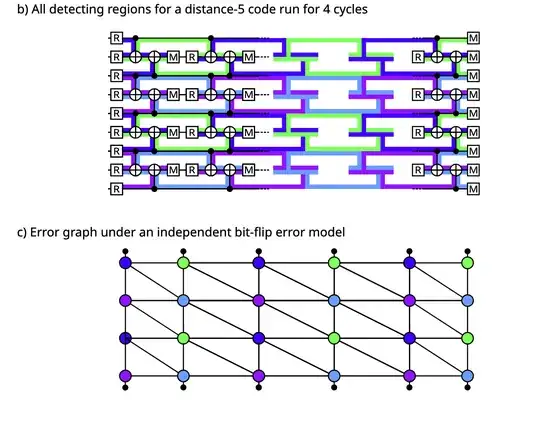
My understanding of decoding in quantum error correction is as follows. Assume you have a code where a single data or ancilla error triggers two detectors (except at space and time boundaries).
Make a circuit composed of stabilizer measurements.
Back propagate any error that anticommutes with the measurement through the (Clifford) circuit as far back as possible before it hits some kind of reset operation.
From this, you get detecting regions. In the bulk, each part of the circuit contains exactly two detecting regions. On the boundaries, you have that each part of the circuit contains only one detecting region.
A detector error model consists of the following: Each detector is a node and the weight of an edge connecting two detectors depends on the probability of an error happening in their overlap region.
Finally, when we run the circuit, the MWPM decoder sees which nodes have lit up (they will be in pairs due to 3, except at the boundary) and it joins edges between them such that the total edge weight is minimized.
This is shown in this paper
My questions is the following and are probably a result of gaps in my understanding regarding both error correction and probability theory:
If two detectors are lit, then there are usually many paths (each corresponding to a different set of errors) that connect them. Do we care about this at all?
If yes, how do we compute the list of all possible error paths from the edge weights we computed in Step 4? If two nodes are lit in the second figure, I need to sum over many paths that connect them? How do I enumerate over all these paths and compute each path's weight?
Why do I sum over all paths if my output is going to be one specific error path (the minimum weight path) that results in the observed syndrome? Isn't the decoder's problem simply to find one set of edges such that the syndrome is explained by it and the weight of that sum is minimized?