I have used an ab initio electronic structure code to calculate the harmonic Raman spectrum of a (molecular) crystal. However, no use of symmetry has been made, and it might well be that some of the vibrational modes that I obtained are actually Raman-inactive. As a matter of fact, when I consider a single cell, two of the vibrational modes I obtain have a high Raman intensity, but when I double the size of the cell, these two modes seem to fuse into a single mode of much lower intensity (not zero, but...). My question(s) would thus be the following.
- Looking at a specific calculated vibrational mode, how can I say whether it is Raman-active or not ? I think I first have to determine the symmetry of my mode, and see if it has the same symmetry as one component of the polarizability tensor, but I'm not sure how to do this, as I am a bit confused with point/space groups. Maybe there is also another way.
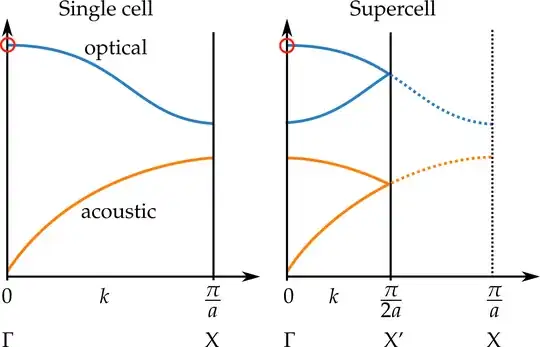
- Looking at a specific calculated vibrational mode in a single cell, can I determine what this mode would look like for the double-sized cell ?
Some more information:
- My system contains 80 atoms (4 molecules) per unit cell, and its space group is P2$_1$/a.
- The two modes I'm talking about are methyl group rotations.
The information about the vibrational modes is stored as an xyz file, containing, in order, the atom type, the position, and the eigenvector (basically, a vector "sits" on every atom). The file would look like this:
80 stable frequency at 155.045 1/cm Raman int. is 1.6100E+02 Ang^4/amu; red. mass is 1.807 a.m.u.; force const. is 0.026 mDyne/Ang. C 8.3725 5.4769 9.6155 -0.0071 0.0657 -0.0516 H 8.2818 6.5453 9.3705 0.2118 0.0540 -0.1846 H 7.6925 5.2982 10.4580 -0.1575 0.2755 -0.1301 H 9.4066 5.2741 9.9201 -0.0804 -0.0702 0.1096 ... C 6.3734 10.0506 7.9586 0.0071 0.0657 0.0516 H 5.3393 9.8479 7.6541 0.0804 -0.0703 -0.1096 H 7.0533 9.8719 7.1161 0.1575 0.2755 0.1301 H 6.4642 11.1191 8.2036 -0.2118 0.0540 0.1846 ...
Thank you in advance!