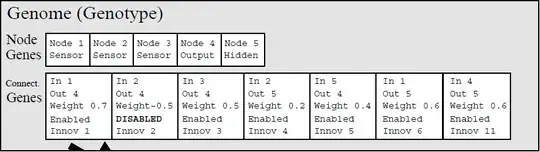
In the original paper the innovation ID is on the connections only.
The connection is the object that is keeping the information; nodes can be discerned by the connections.
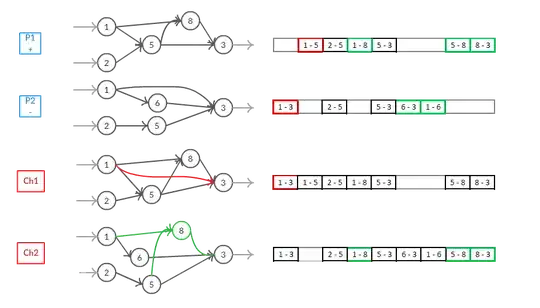
This image represents a possible crossover operator that makes a distinction between disjoint and excess genes and therefore creates children depending on these. It's part of my master thesis, I'd be glad to expand the topic, but for now I'll just use it as example.
In the image we assume that the connections that are joining two nodes have the same innovation number.
As you can see, there is no need of assigning an innovation number to the nodes: nodes are just a result of what the connection say. This also allows for a more dynamic approach that can be used to spot invalid nets even before building them (checking if there are cyclics or nodes that aren't receiving any input or giving any output) and correct them in order to obtain only valid graphs.
Nodes are added just because there is a connection that is pointing to that specific node. This is enough to grant its presence (node number 8 in child 2).
As last point, for the data normalization theory (Normalization is a process of organizing the data in database to avoid data redundancy, insertion anomaly, update anomaly & deletion anomaly) we should avoid rendundancy at any cost and this is why we should try to keep track of the smallest amount of object possibles. So if we can deduct the nodes from the connections we should do it.