I'm not very knowledgeable in this field but I'm wondering if any research or information already exists for the following situation:
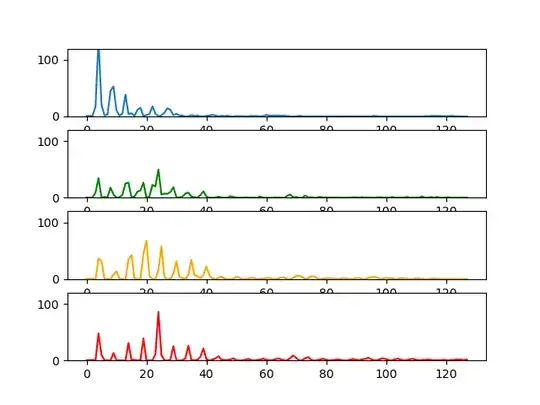
I have some data that may or may not look similar to each other. Each represents a node that represents a vector of size 128.
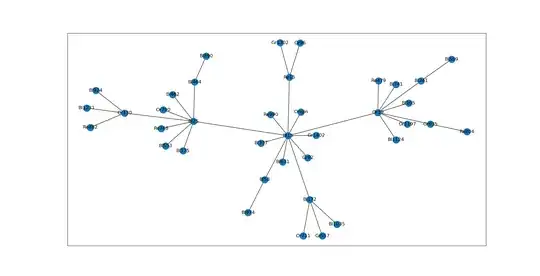
And they are inserted into a tree graph according to similarity.
and a new node is created with an edge connecting the most similar vertex node found in the entire tree graph.
Except I'm wasting a lot of time searching through the entire graph to insert each new node when I could narrow down my search according to previous information. Imagine a trunk node saying "Oh, I saw a node like you before, it went down this branch. And don't bother going down that other branch." I could reduce the cost of searching the entire tree structure if there was a clever way to remember if a similar node went down a certain path.
I've thought about some ways to use caching or creating a look-up table but these are very memory intensive methods and will become slower the longer the program runs on. I have some other ideas I am playing around with but I was hoping someone could point me in the right direction before I started trying out weird ideas.
Edit: added a better (more realistic) graph picture